Single-Cell Microbial WGS kit high-quality microbial genome recovery at scale

Single-Cell Microbial WGS kit barcodes up to 10,000 microbial single amplified genomes (SAGs) per experiment, enabling genome recovery for individual cells after sequencing. Start by encapsulating microbial cells from suspension into semi-permeable capsules (SPCs), then seamlessly proceed with lysis, genome amplification, barcoding and library preparation. Optimized for Illumina short-read sequencing, the kit can also be adapted to any commercial NGS platform, including short- and long-read sequencing.
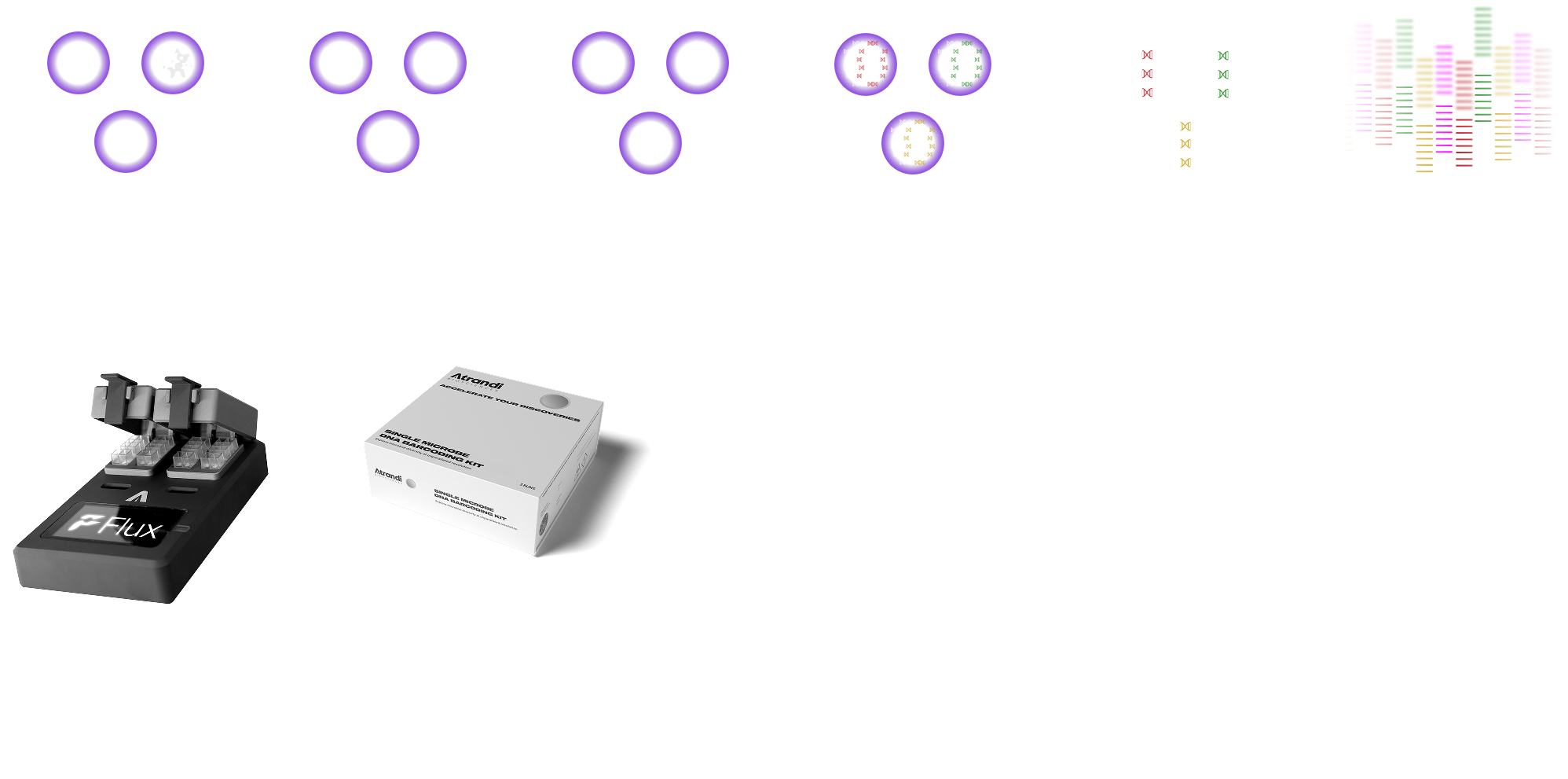
Features




Publications Using our Single-Cell Microbial WGS Kit

Kit components

- 2x Lysis Buffer
- Neutralization Buffer
- 50x Wash Buffer
- 10x Stop Buffer
- Water
- Debranching Enzyme
- 10x Debranching Buffer
- End Prep Enzyme Mix
- End Prep Reaction Buffer
- WGA Polymerase
- WGA Enhancer
- 10x WGA Reaction Buffer
- dNTP Mix
- Primer Mix
- Fragmentation Enzyme Mix
- Fragmentation Reaction Buffer
- Ligation Adapter
- Ligation Master Mix
- Ligation Enhancer
- Amplification Master Mix
- Indexing Primer Mix
Specifications
| Catalog number | CKP-BARK8 |
| Sufficient for | 2 runs x 10,000 barcoded microbial cells |
| Sample type | Microbial cell suspension |
| For use with (equipment) |
|
| Sequencing | Illumina MiSeq, NextSeq 550, NovaSeq |
| Shipping conditions | Dry Ice |
| Storage | -20 °C |
| Intended Use | Research Use Only |
SAMPLE TYPES
Designed for versatility, our technology seamlessly integrates with diverse microbiomes and isolates, provided that the cells are in aqueous suspension and free from large particulates (>40 μm).




Lysis
SPCs can withstand harsh lysis conditions, which can be carried out in multiple steps. While the kit includes an alkaline lysis buffer, researchers have the flexibility to use their preferred lysis method, with the exception of mechanical disruption.



MULTIPLEXING CAPABILITIES
Samples can be multiplexed at various stages of the workflow, maximizing efficiency.
Applications




PROOF OF CONCEPT SEQUENCING SERVICE
Thinking about doing single-cell microbial genomics but not sure whether Single-Microbe DNA Barcoding kit will be a good fit?
Test your sample's full potential quickly and easily by sending it to us. The proof of concept experiment includes the following:
- Sample preparation
- Cell encapsulation into SPCs
- Single-microbe DNA barcoding
- Sequencing on the Illumina platform (NextSeq 550 or NovaSeq)
- Data demultiplexing by cellular barcodes

ADVANCED DATA ANALYSIS
We've partnered with the Single Cell Genomics Center at Bigelow Laboratory to offer advanced bioinformatics services for microbiologists lacking data analysis resources
Learn more